Welcome to the Genomic Annotation Gathering database
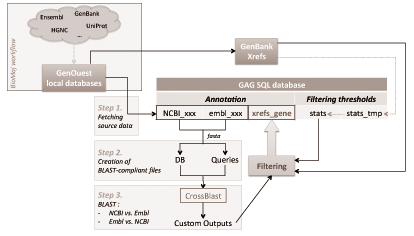
The Genomic Annotation Gathering tool is a database which aims to provide an enriched cross-references database to scientific community. Cross-references are achieved by using sequence similarity comparison between transcripts originating from GenBank and Ensembl, a custom filtering process, and posterior comparison with official cross-reference tables provided by GenBank. This way, we are able to propose new (predicted) cross-references that were unavailable before, to complete known (official) data.
Available annotation data includes all transcripts and their identifiers, functionnal description of genes, chromosomal localization, gene symbols, gene homologs for model species (human, chicken, mouse), and several identifiers to link those genes to external databases (UniProt, HGNC).
Queries should be made either by chromosomal localization (to return all known genes and their cross-references between two loci), gene symbol (to return both GenBank and Ensembl identifiers that link to this gene), or unique gene identifier.
An example query "14:31066124-31082030" will search in gag for chromosome 14 on specified location
If you use GAG, please cite:
Obadia T., Sallou O., Ouedraogo M., Guernec G. and Lecerf F. The GAG database: A new resource to gather genomic annotation cross-references. Gene (2013) pp.http://dx.doi.org/10.1016/j.gene.2013.06.063
Copyright © 2005-2012 Biogenouest® -
Legal notice -
Contact : ![]()